Data Analysis
We offer comprehensive data analysis services to explore both the data we produce with our platforms and the data from your own experiments. Our bioinformatics team will guide you in understanding your results with tables, high-resolution plots, and a clear pdf report describing the analytical steps.
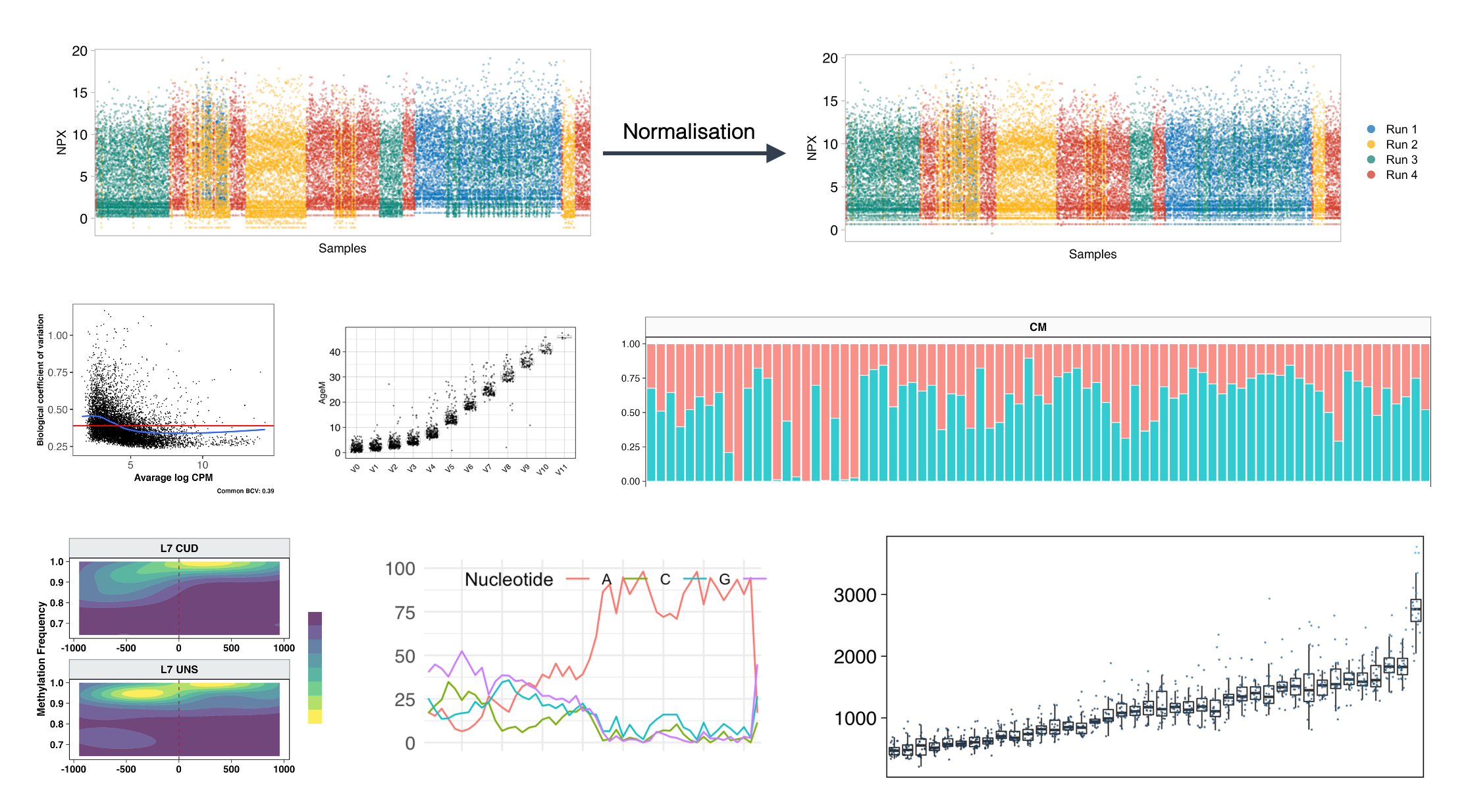
Raw data analysis and quality check
Each platform produces raw data that need to be processed, normalised, and checked before any type of downstream analysis. For example, regarding RNA sequencing, this step consists of basecalling, reads quality check and filtering, alignment against reference transcriptome, reads count, and normalisation.
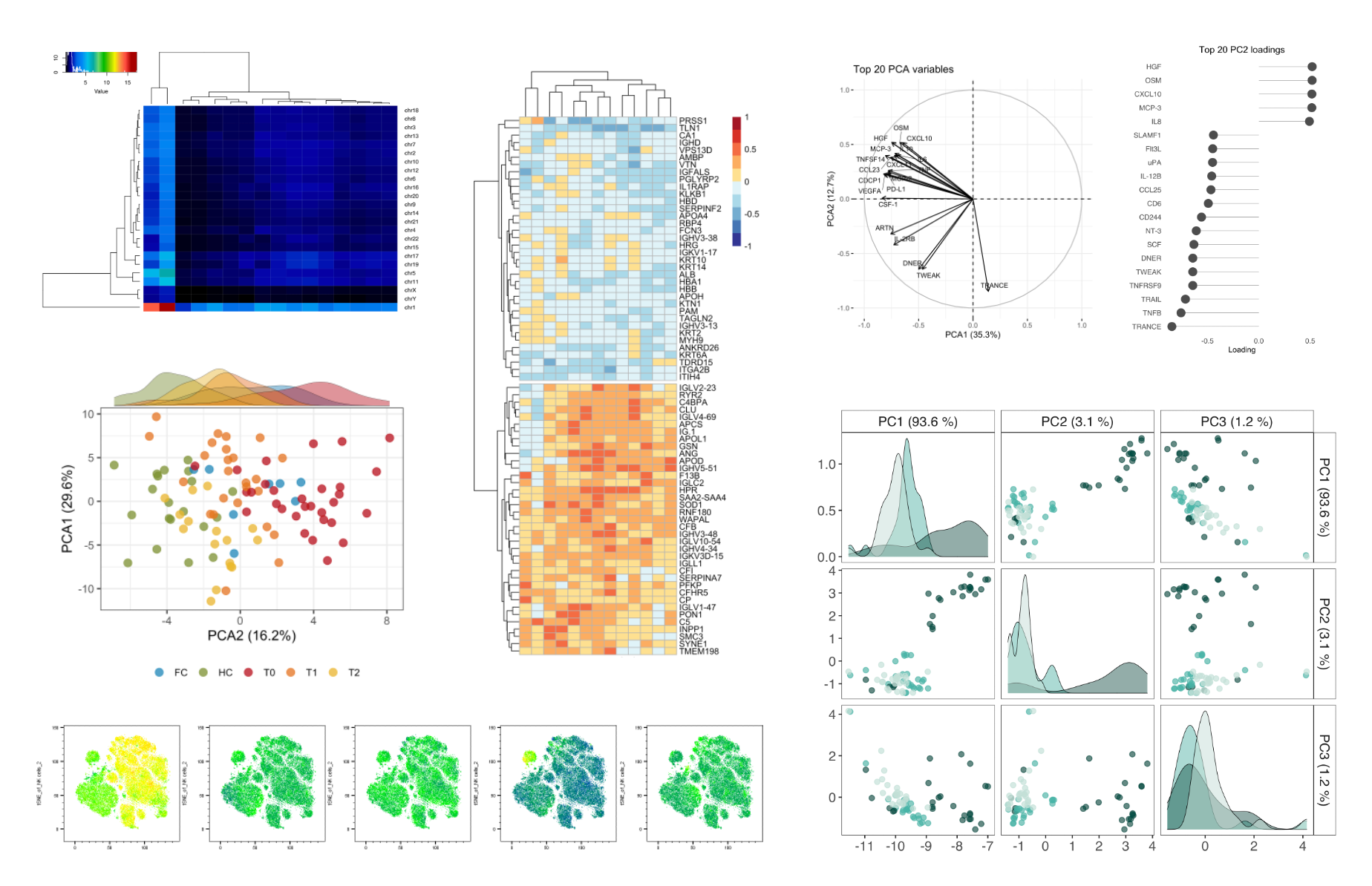
Exploratory data analysis
Dimensionality reduction techniques, such as PCA, T-SNE, UMAP, MOFA, or unsupervised hierarchical clustering strategies are commonly applied to get an overview of large biological datasets. These approaches allow to highlight sample similarities driven by the whole data set(s).
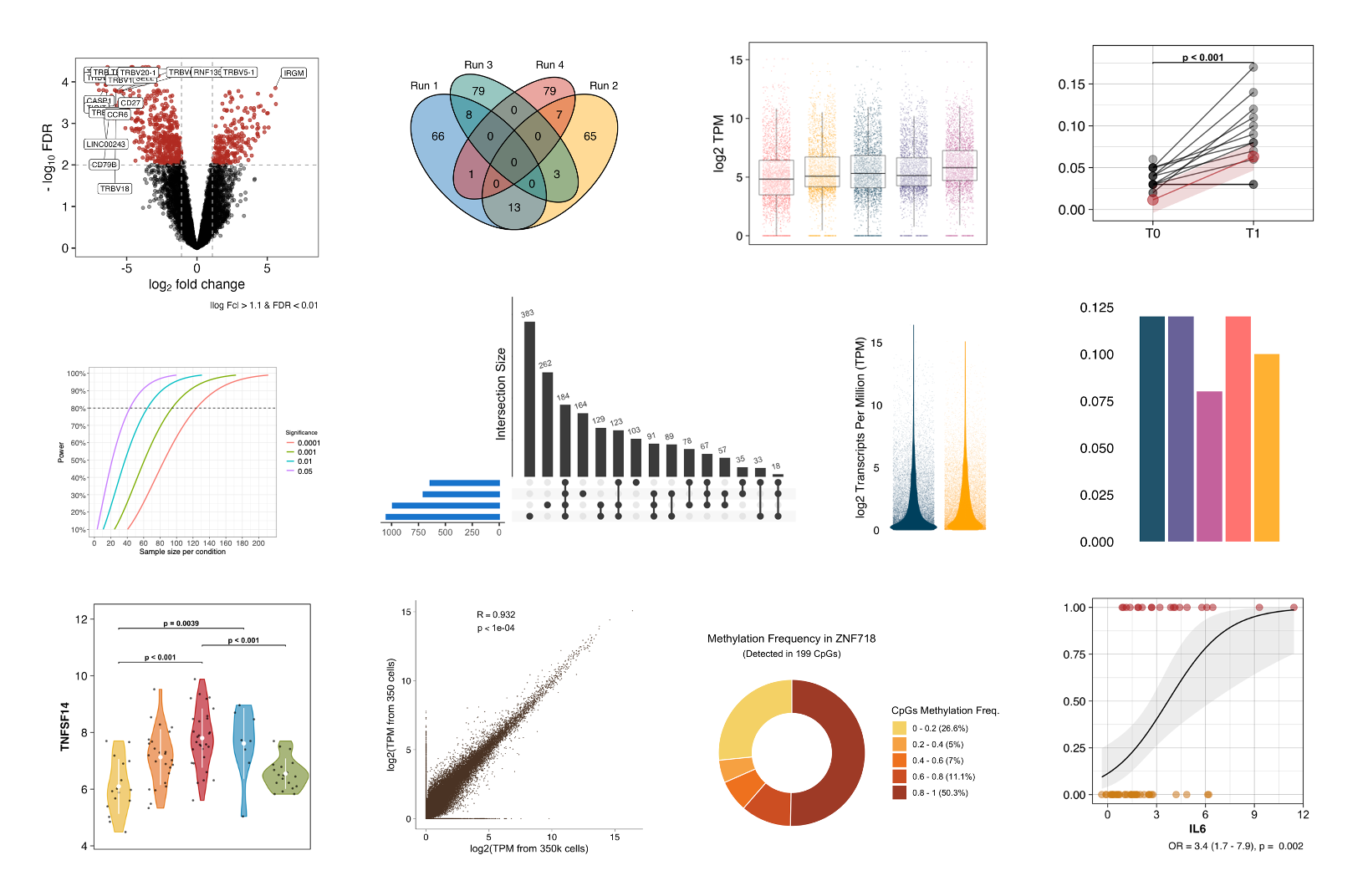
Statistical analysis
The main objective of any experimental project is to summarise, describe, and visualise the data, measure relationships within the data, and test hypotheses using statistical tests. Our pipelines are designed to offer a reproducible workflow and are able to choose the right test depending on the number of comparisons, the normality, and the homogeneity of the data distributions.
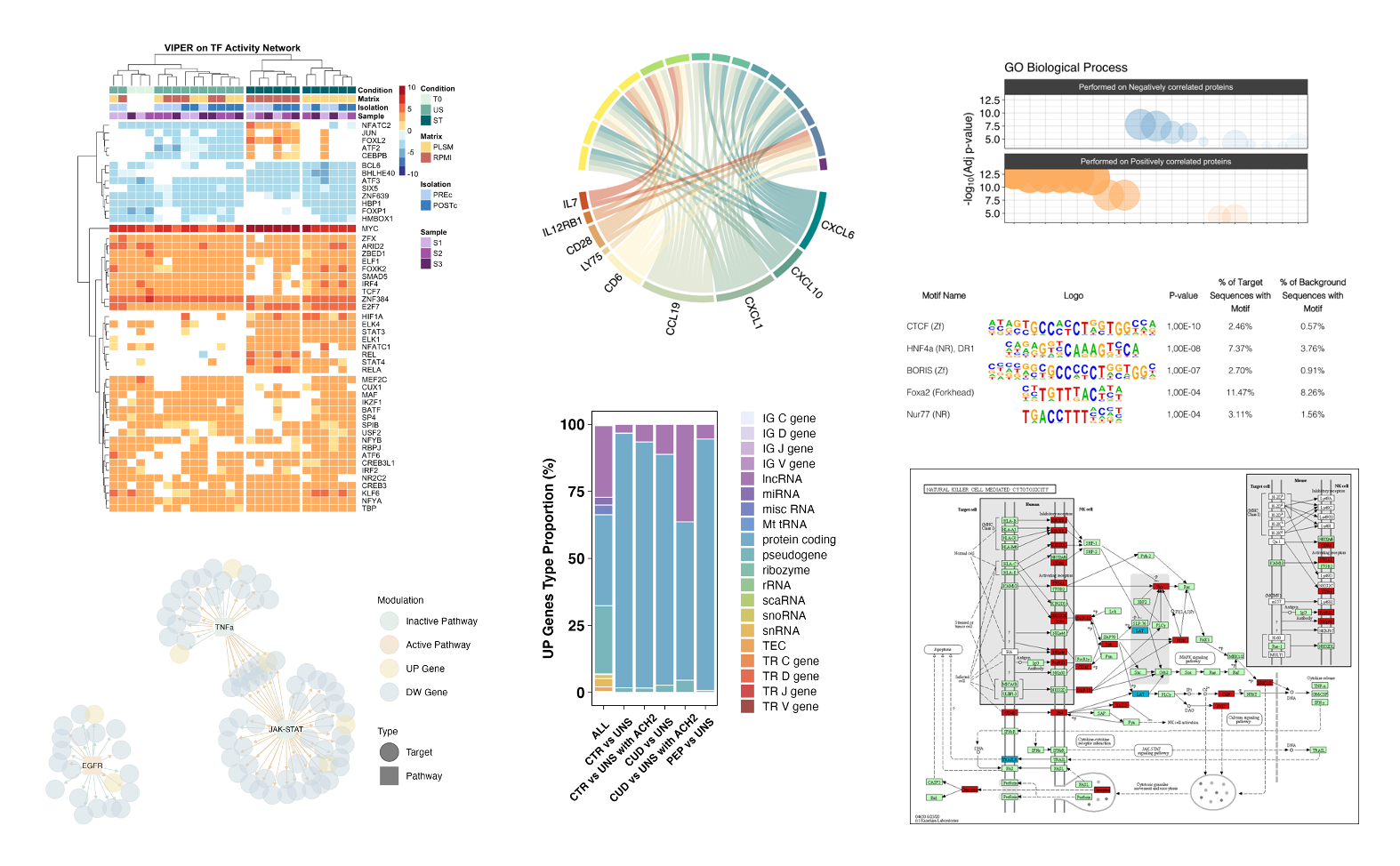
Functional analysis
Hypothesis tests applied to omics data generally produce a list of features that are significantly different between conditions. These features can be, for example, transcripts, proteins, and methylation sites. Functional analyses provide an overview of the biological roles of these deregulated features. Depending on the data type and the scientific questions, we perform gene ontology (GO), Gene Set Enrichment Analysis (GSEA), and Virtual Inference of Protein-activity by Enriched Regulon analysis (VIPER), for instance, useful to infer the transcription factors' activity starting from RNA-seq data.
Multi-omics data integration
A single omics layer is a wealth of information, but by integrating information from multiple layers, researchers gain an unprecedented depth of insight into the intricate web of interactions within biological systems. Simplifying, this type of analysis consists of building a network where each node is a feature of any omics layer and the nodes are connected using both public information and relationships that came out from the data.
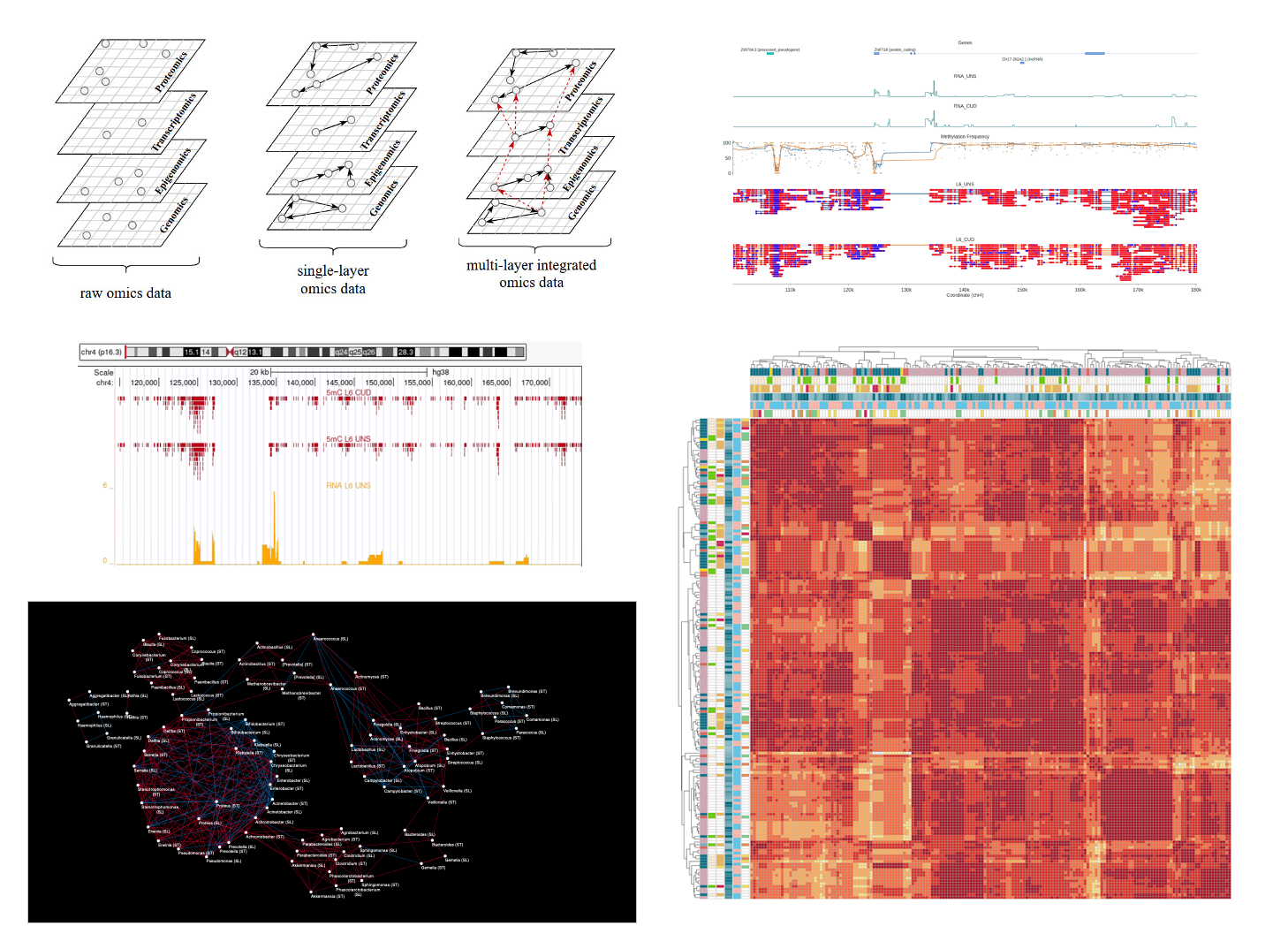
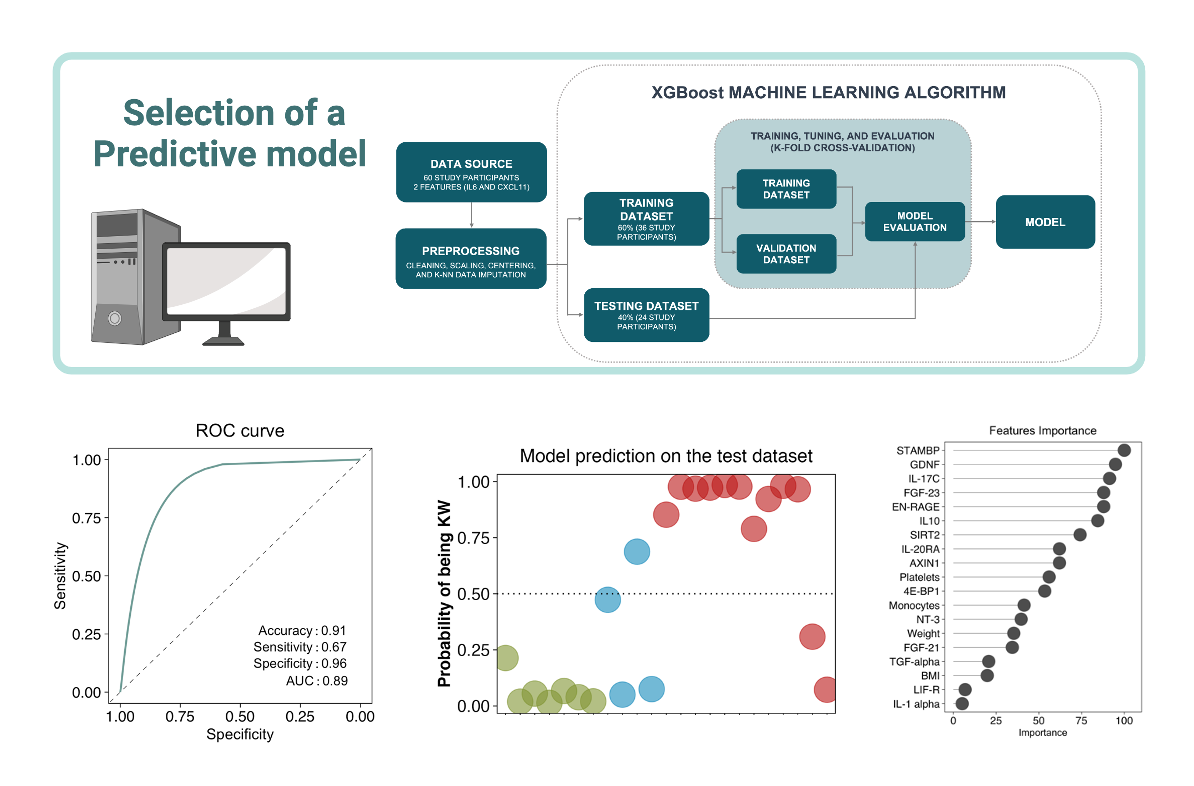
Artificial Intelligence models development
We provide the possibility to build supervised, and semi-supervised Artificial Intelligence models to predict any feature you are interested in, given the dataset you choose to provide to us. One example is using as the predictors metrics from multi-omics profiles, and as the feature to predict the responsiveness or the adverse effects after inoculation of specific therapies. Predictive models can be trained to determine which outcome is expected, and what individual characteristics promote one outcome instead of another. This kind of approach is powerful to discern whether a specific therapeutic strategy will be effective on a specific individual, and also for the discovery of biomarkers that are informative for the discrimination between biological conditions.
Mathematical Modelling
Systems biology has evolved during the last decades to provide progressively more insightful mechanistic models of biological systems. Complex hypotheses on system behaviour can be tested through the development of a mathematical model, merging previously defined knowledge with novel insights given by the most recent experiments. We offer the opportunity to test the rules you think dictate the relationship between the elements of your biological system (hypotheses) to determine to what degree they explain the observed data. This process is subdivided into (i) translation of your hypotheses in mathematical form to build the mathematical model, and (ii) model fitting and validation against your experimental data.
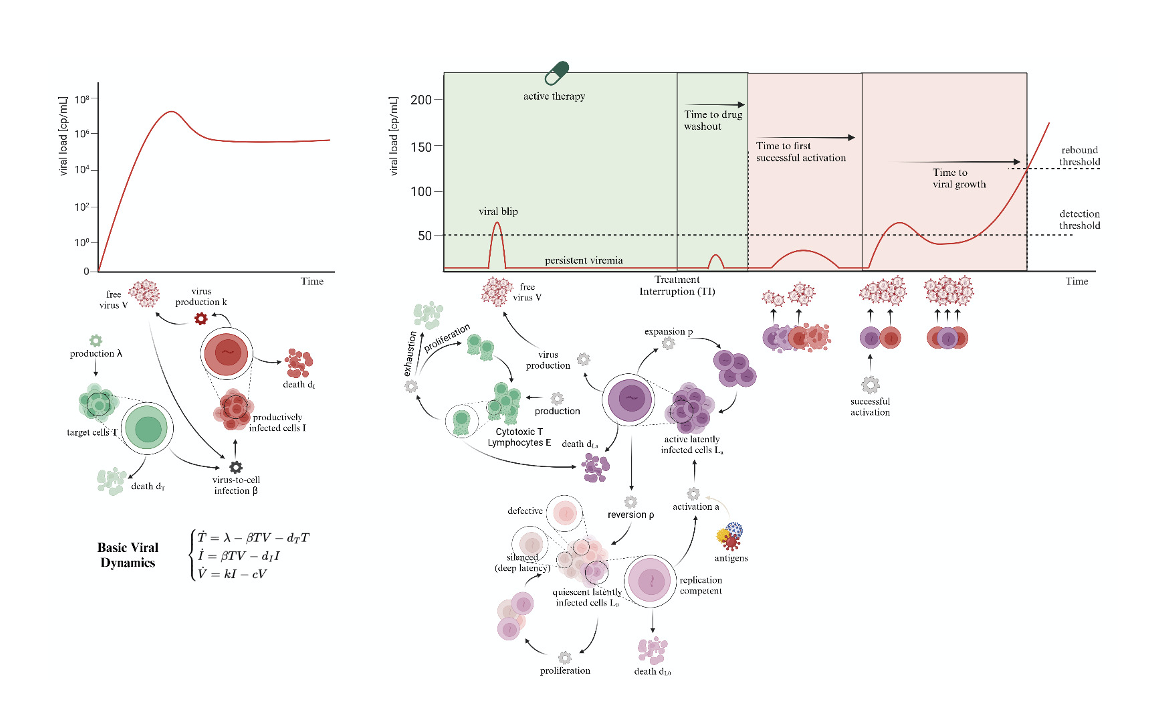
Data interpretation
Every data analysis that we offer is accompanied by a detailed explanatory report. However, if you also prefer to have help interpreting the results from a biological point of view, our team of doctors, biologists, bioinformaticians, and bioengineers can offer this service.
All plots in this section were created by the Probiomics team.